Antrodia cinnamomea exerts an anti-hepatoma effect by targeting PI3K/AKT-mediated cell cycle progression in vitro and in vivo
Abstract
Antrodia cinnamomea is extensively used as a traditional medicine to prevention and treatment of liver cancer. However, its comprehensive chemical fingerprint is uncertain, and the mechanisms, especially the potential therapeutic target for anti-hepatocellular carcinoma (HCC) are still unclear. Using UPLC‒Q-TOF/MS, 139 chemical components were identified in A. cinnamomea dropping pills (ACDPs). Based on these chemical components, network pharmacology demonstrated that the targets of active components were significantly enriched in the pathways in cancer, which were closely related with cell proliferation regulation. Next, HCC data was downloaded from Gene Expression Omnibus database (GEO). The Cancer Genome Atlas (TCGA) and DisGeNET were analyzed by bioinformatics, and 79 biomarkers were obtained. Furtherly, nine targets of ACDP active components were revealed, and they were significantly enriched in PI3K/AKT and cell cycle signaling pathways. The affinity between these targets and their corresponding active ingredients was predicted by molecular docking. Finally, in vivo and in vitro experiments showed that ACDPs could reduce the activity of PI3K/AKT signaling pathway and downregulate the expression of cell cycle-related proteins, contributing to the decreased growth of liver cancer. Altogether, PI3K/AKT-cell cycle appears as the significant central node in anti-liver cancer of A. Cinnamomea.
Figures 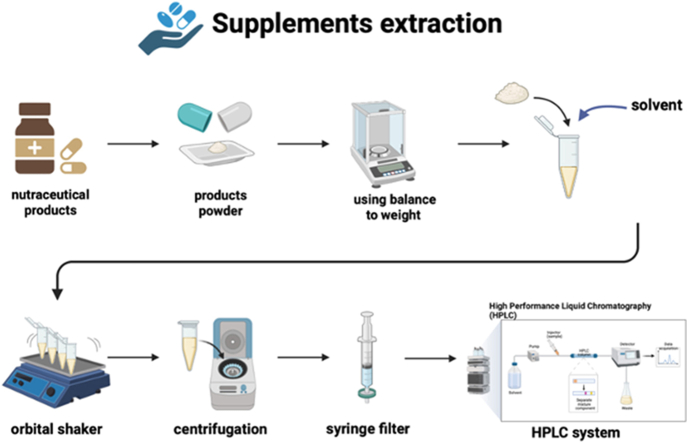
Graphical abstract
Graphical abstract
Graphical abstract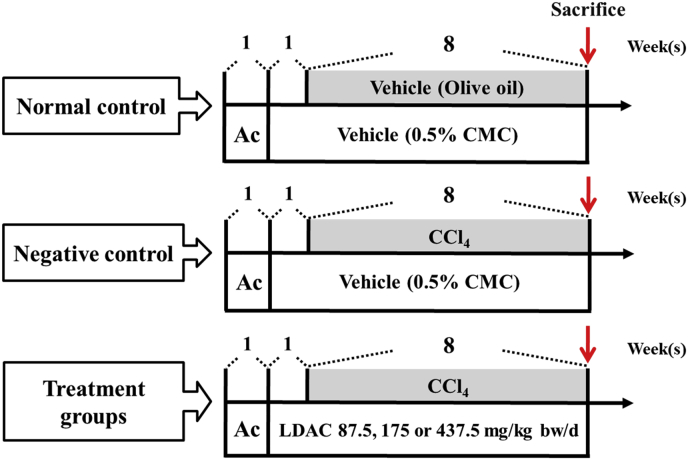
Figure 1
Flow chart of the prediction…
Figure 1
Flow chart of the prediction for targets of active components from ACDPs by…
Figure 1 Flow chart of the prediction for targets of active components from ACDPs by bioinformatics.
Figure 2
Identification result of chemical composition.…
Figure 2
Identification result of chemical composition. (A) Mass spectra and fragmentation pathways of antcin…
Figure 2 Identification result of chemical composition. (A) Mass spectra and fragmentation pathways of antcin A. (B) Sciex OS identification interface of antcin A. (C) Mass spectra and fragmentation pathways of antcin K.
Figure 3
Results of targets of active…
Figure 3
Results of targets of active components from ACDPs. (A) Component-target network diagram of…
Figure 3 Results of targets of active components from ACDPs. (A) Component-target network diagram of ACDPs. (B) Histogram of GO enrichment analysis of targets. (C) Bubble chart of KEGG enrichment analysis of targets.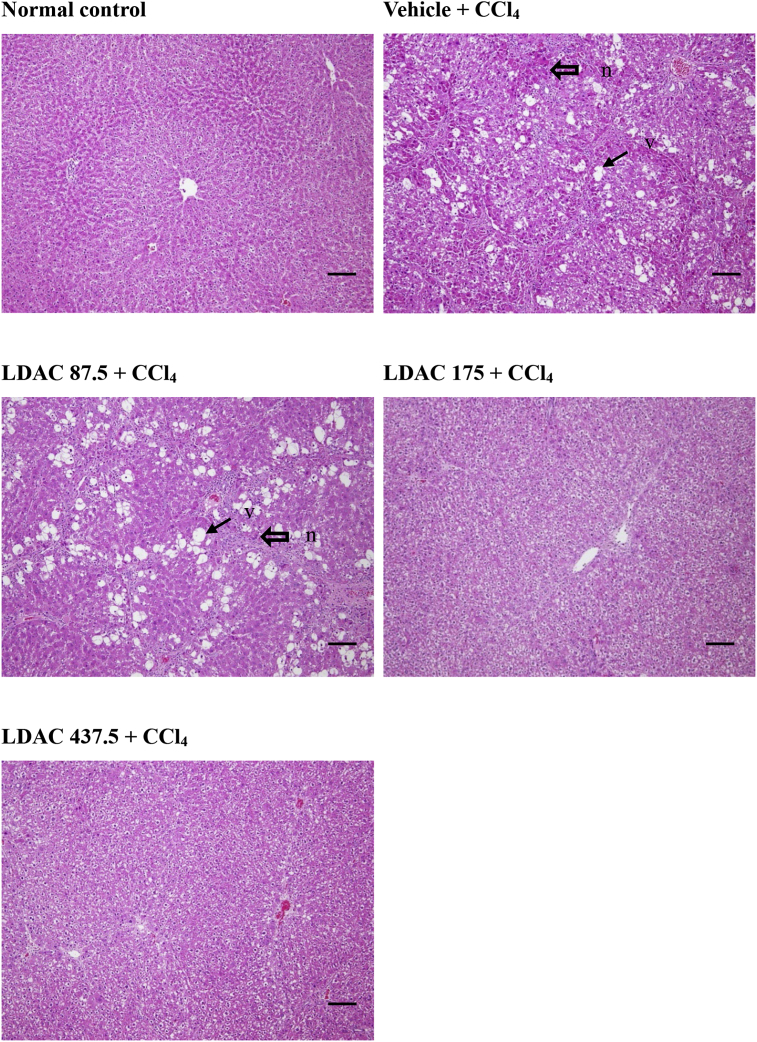
Figure 4
Results of DEGs in HCC…
Figure 4
Results of DEGs in HCC related datasets in GEO database. (A)‒(D) PCA diagram…
Figure 4 Results of DEGs in HCC related datasets in GEO database. (A)‒(D) PCA diagram of GSE14520, GSE45267, GSE64041, and GSE112790. (E)‒(H) Volcano map of DEGs in GSE14520, GSE45267, GSE64041, and GSE112790. (I)‒(L) Heat map of DEGs in GSE14520, GSE45267, GSE64041, and GSE112790. (M) Heat map of TOP 20 up-regulated and down-regulated DEGs.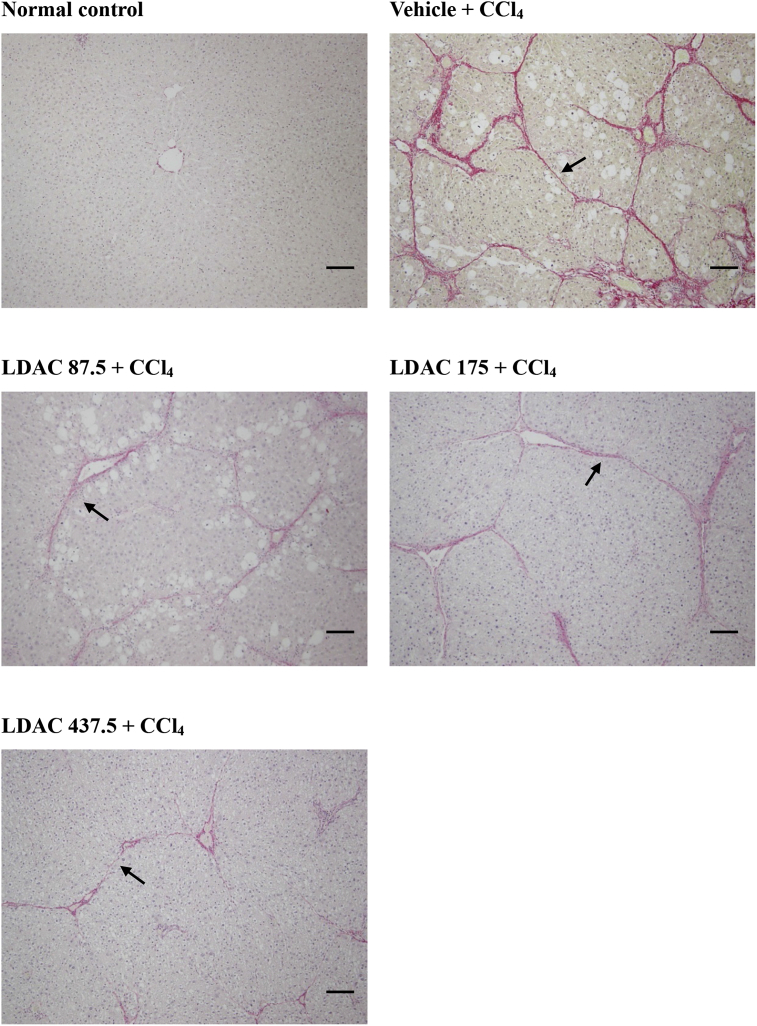
Figure 5
Results of DEGs filtered and…
Figure 5
Results of DEGs filtered and selected by TCGA database. (A)‒(E) Survival curves of…
Figure 5 Results of DEGs filtered and selected by TCGA database. (A)‒(E) Survival curves of AURKA, CCNB1, CDK1, SAE1 and TOP2A in HCC patients. (F)‒(J) ROC curves of AURKA, CCNB1, CDK1, SAE1 and TOP2A in HCC patients.
Figure 6
Results of enrichment analysis of…
Figure 6
Results of enrichment analysis of the HCC-related targets of ACDPs. (A) Veen map…
Figure 6 Results of enrichment analysis of the HCC-related targets of ACDPs. (A) Veen map of active ingredients and HCC targets. (B) Circos diagram of KEGG pathway enrichment analysis of the nine common targets. (C) Network of the nine common targets and ACDPs active ingredients. (D) Three-dimensional binding mode and two-dimensional binding mode of zhankuic acid E and PIK3CA (PDB: 6PYS). (E) Three- and two-dimensional binding mode of ganoderiol B and CDK1 (PDB: 4YC3).
Figure 7
ACDPs repress the proliferation of…
Figure 7
ACDPs repress the proliferation of HCC cells in vitro through PI3K/AKT/cell cycle axis.…
Figure 7 ACDPs repress the proliferation of HCC cells in vitro through PI3K/AKT/cell cycle axis. (A)‒(C) CCK-8 for the viability of HCC cells, cultured in medium containing 0, 0.4, 0.8, 1.6, 3.2 mg/mL ACDPs for 24 and 48 h. Data are presented as mean ± SD, ∗P < 0.05, ∗∗P < 0.01 and ∗∗∗P < 0.001 compared to control group. (D) Colony formation experiments for the effect of ACDPs on the proliferation of HCC cells (n = 6). (E)‒(G) EdU assay for the proliferation of HCC cells treated with the 3.2 mg/mL ACDPs or DMSO (control) for 24 h. Bar = 25 μm. (H) Western blot and densitometric analysis of expression of cyclin E1, CDK1, cleaved-caspase 8, PIK3CA, p-AKT1 and AKT1 in Huh-7 cells, cultured in medium containing 3.2 mg/mL ACDPs or DMSO for 24 h ∗P < 0.05, ∗∗∗P < 0.001 compared to control group.
Figure 8
ACDPs inhibited tumor growth and…
Figure 8
ACDPs inhibited tumor growth and downregulated the expression of cell cycle-related protein in…
Figure 8 ACDPs inhibited tumor growth and downregulated the expression of cell cycle-related protein in HCC-xenograft mice. (A) and (B) The images of mice in each group and their corresponding transplanted tumors. (C) The effect of ACDPs on tumor weight of HCC-allograft mice (n = 5). ∗P < 0.05, ∗∗P < 0.01 compared to model group. (D) The effect of ACDPs on tumor volume of HCC-allograft mice (n = 5). ∗P < 0.05, ∗∗P < 0.01 and ∗∗∗P < 0.001 compared to model group. (E) Immunohistochemistry analysis for the expression of cyclin E1, CDK1, cleaved-caspase 8, PIK3CA, p-AKT1 and AKT1 in tumor tissues from model and ACDPs group of HCC-allograft mice (n = 5). ∗P < 0.05, ∗∗P < 0.01 and ∗∗∗P < 0.001 compared to model group. Bar = 50 μm. All figures (9)