The complete mitochondrial genome of the edible mushroom Grifola frondosa
Abstract
The culinary-medicinal mushroom Grifola frondosa is widely cultivated in East Asia. In this study, the complete mitochondrial genome of G. frondosa was determined using Illumina sequencing. The circular molecule was 197,486 bp in length with a content of 25.01% GC, which was one of the largest mitochondrial genomes in the order Polyporales. A total of 39 known genes encoding 13 common mitochondrial genes, 24 tRNA genes, 1 ribosomal protein s3 gene (rps3), and 1 DNA polymerase gene (dpo) were predicted in this genome. The phylogenetic analysis showed that G. frondosa clustered together with Sparassis crispa, Laetiporus sulphureus, Wolfiporia cocos, and Taiwanofungus camphoratus. The complete mitochondrial genome reported here may provide new insight into genetic information and evolution for further studies.
Figures 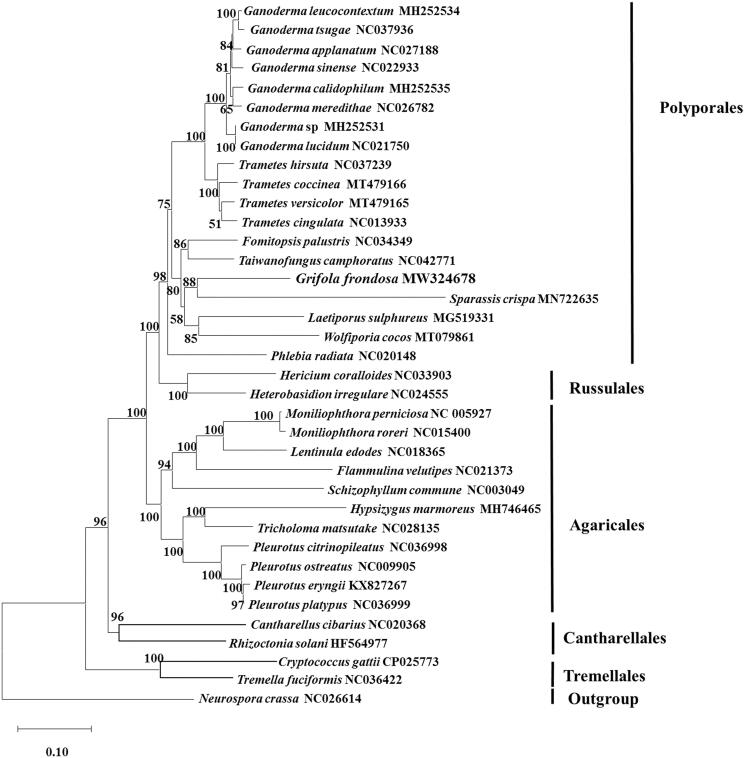
Figure 1.
Phylogenetic analysis of 35 species…
Figure 1.
Phylogenetic analysis of 35 species of Basidiomycota conducted by neighbor-joining method based on…
Figure 1. Phylogenetic analysis of 35 species of Basidiomycota conducted by neighbor-joining method based on concatenated amino acid sequences of 13 conserved protein-coding genes (common in all the species), including atp6, atp8, atp9, cob, cox1, cox2, cox3, nad1, nad2, nad3, nad4, nad5, and nad6. Neurospora crassa (NC_026614) was served as outgroup. The bootstrap support values were shown at each node.