MicroRNA-like small RNAs prediction in the development of Antrodia cinnamomea
Figures
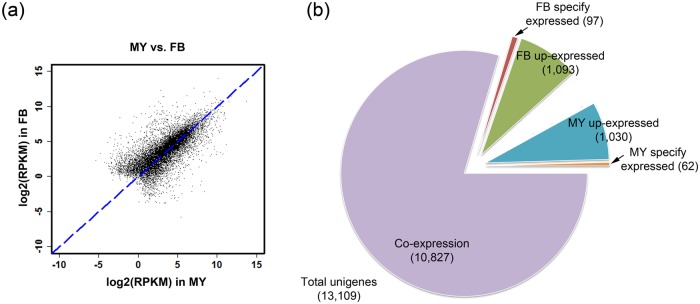
Fig 1. Differentially expressed gene analysis of…
Fig 1. Differentially expressed gene analysis of wild-type fruiting bodies and liquid cultured mycelium of…
Fig 1. Differentially expressed gene analysis of wild-type fruiting bodies and liquid cultured mycelium of A. cinnamomea. F: wild-type fruiting bodies; M: liquid cultured mycelium. (a) Scatter plot of unigenes from A. cinnamomea RNA-seq. (b) Pie chart of the DEG distribution in wild-type fruiting bodies and liquid cultured mycelium. FDR <0.05 and fold change ≥2 or ≤0.5 were defined as differential expression.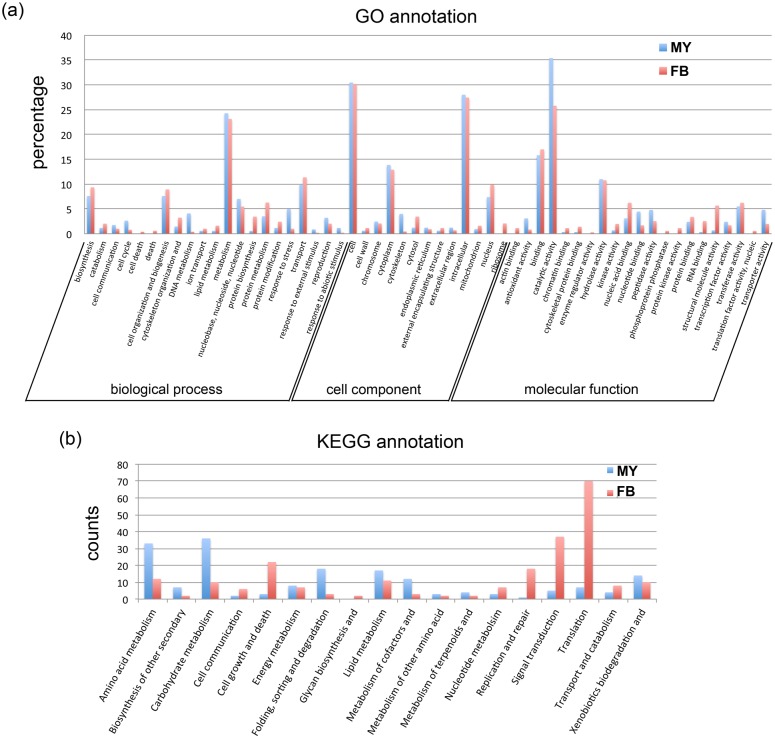
Fig 2. Gene ontology classification and KEGG…
Fig 2. Gene ontology classification and KEGG annotation of DEGs between wild-type fruiting body and…
Fig 2. Gene ontology classification and KEGG annotation of DEGs between wild-type fruiting body and liquid cultured mycelium. MY: unigenes upregulated in liquid cultured mycelium, FB: unigenes upregulated in wild-type fruiting bodies. (a) GO annotation. 2,282 unigenes from DEGs were analyzed with blast2GO to obtain the GO terms. And the GO term were classified with CateGOrizer and separated into three major categories. (b) KEGG annotation. 2,282 unigenes from DEGs were submitted to KAAS to get the KEGG metabolic pathway classification.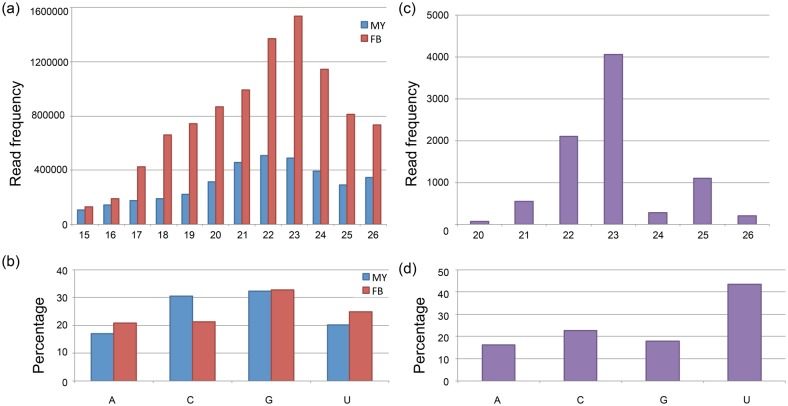
Fig 3. General features of the sRNAs…
Fig 3. General features of the sRNAs and predicted miRNA in A . cinnamomea .
(a) length…
Fig 3. General features of the sRNAs and predicted miRNA in A. cinnamomea. (a) length distribution of total clean reads of sRNA library from MY and FB, (b) 5′ end nucleotide frequency of sRNAs from MY and FB, (c) length distribution of predicted novel milRNAs, (d) 5′ end nucleotide frequency of predicted novel milRNAs.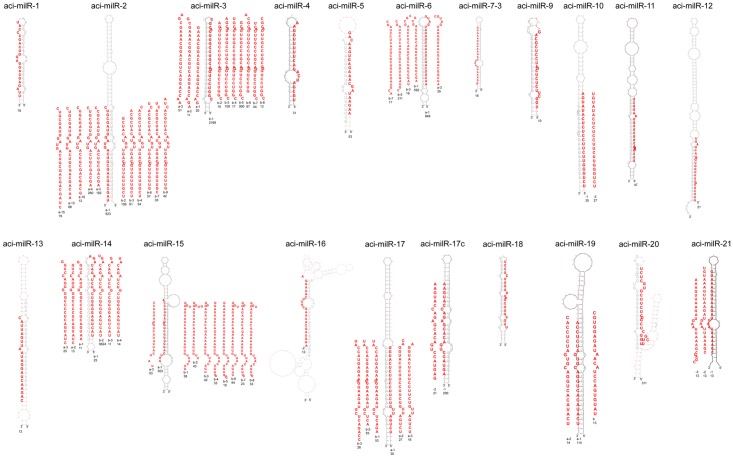
Fig 4. Secondary hairpin structures of milRNA…
Fig 4. Secondary hairpin structures of milRNA from A . cinnamomea predicted by RNAfold.
The milRNAs…
Fig 4. Secondary hairpin structures of milRNA from A. cinnamomea predicted by RNAfold. The milRNAs were sorted by the source of the precursors. milRNAs are marked in bold red and the reads are listed below.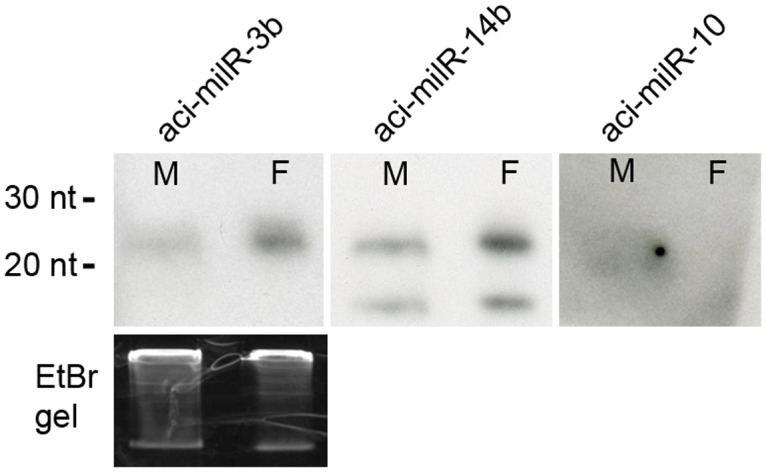
Fig 5. milRNAs identified in this study…
Fig 5. milRNAs identified in this study with Northern blot.
Fig 5. milRNAs identified in this study with Northern blot.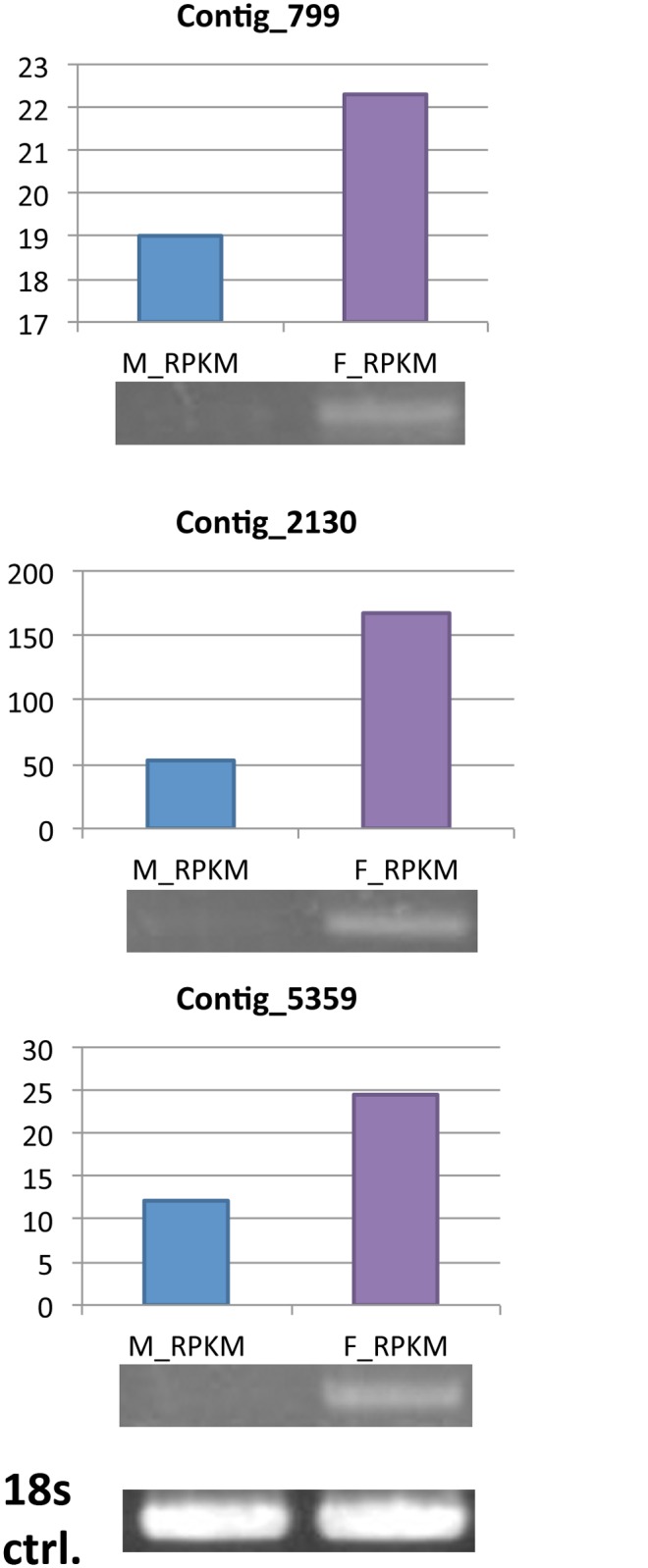
Fig 6. RT-PCR of DCL-2 (Contig_799), QDE-2…
Fig 6. RT-PCR of DCL-2 (Contig_799), QDE-2 (argonaute protein, Contig_2130), DCL-1 (Contig_5359) and 18s rRNA…
Fig 6. RT-PCR of DCL-2 (Contig_799), QDE-2 (argonaute protein, Contig_2130), DCL-1 (Contig_5359) and 18s rRNA genes in A. cinnamomea.